Detection of mecA Gene and Identification of Potential Methicillin-resistant Staphylococcus Aureus in Hospital Wastewater Samples
DOI:
https://doi.org/10.31357/ait.v1i1.4836Abstract
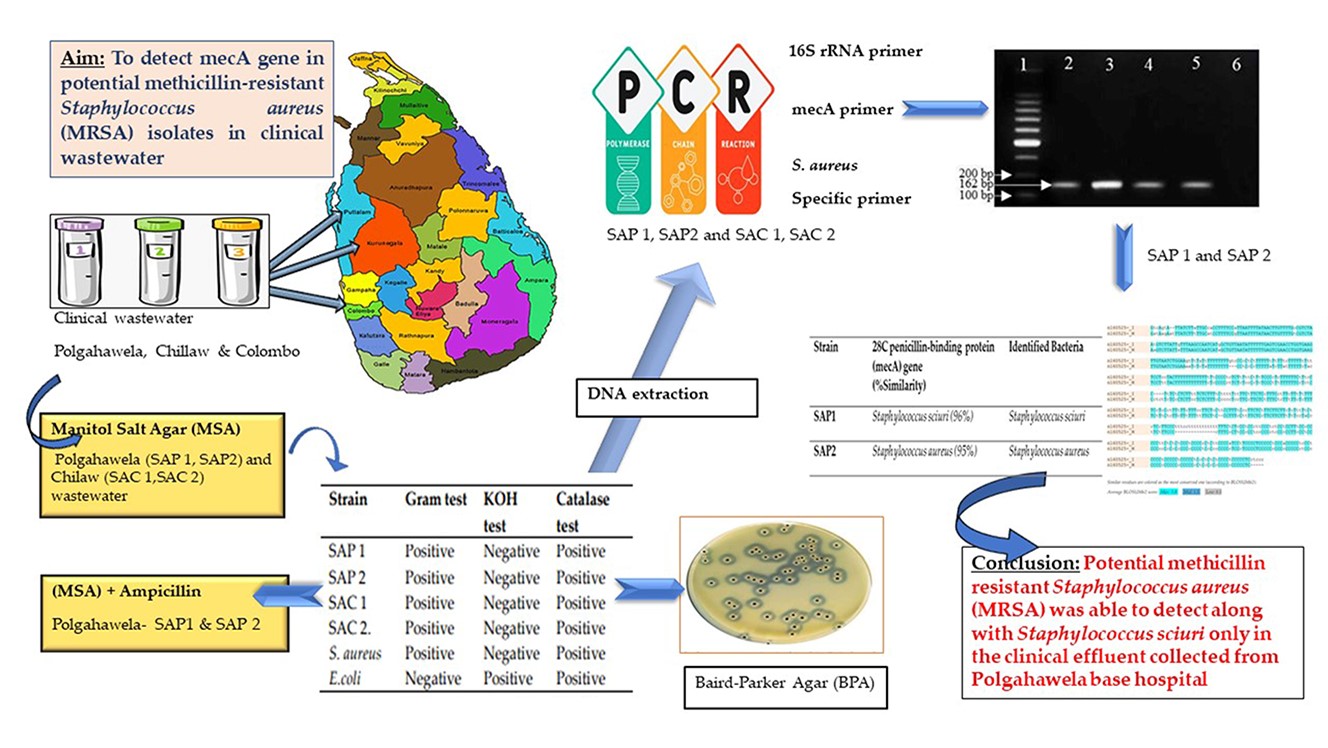
Methicillin-resistant Staphylococcus aureus (MRSA) is one of the most important causes of hospital infections worldwide. Methicillin-resistant S. aureus (MRSA) tends to be resistant to multiple antibiotics. High-level resistance to antibiotics is caused by the mecA gene, which encodes an alternative penicillin-binding protein, PBP 2a. The present study was aimed to detect mecA in potential Methicillin-resistant Staphylococcus aureus (MRSA) isolates in clinical wastewater. Three hospital wastewater samples were collected and the bacteria were isolated in manitol salt agar (MSA) medium. PCR was performed and sequencing was done to screen the clinical bacterial isolates. Antibiotic resistance was also tested at the levels of 50, 100, 200 and 300 μg/mL ampicillin. The sequences were analyzed using BLAST (NCBI) and EMBOSS Needle tool (EMBL-EBI). DNA extracted from these bacterial isolates amplified further with mecA gene specific primers. Randomly selected two bacterial isolates of Polgahawela hospital effluent were able to grow at 200 μg/mL ampicillin. Sequence analysis of amplified mecA gene product of these two bacterial isolates showed sequence similarity with the penicillin-binding protein (mecA) gene of Staphylococcus aureus strain and methicillin-resistance gene region of Staphylococcus sciuri 28C with 95% and 96% identity respectively. Pairwise alignment results proved 89.6% sequence similarity between the two sequences. In conclusion, potential methicillin resistant staphylococcus aureus (MRSA) along with Staphylococcus sciuri was able to detect only in the clinical effluent collected from Polgahawela base hospital.
Downloads
Published
How to Cite
License
Copyright (c) 2021 Kalaivani Vivehananthan, M. P. D. L. Luphzhy

This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License.
The Authors hold the copyright of their manuscripts, and all articles are circulated under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, as long as that the original work is properly cited.
The use of general descriptive names, trade names, trademarks, and so forth in this publication, even if not specifically identified, does not imply that these names are not protected by the relevant laws and regulations. The authors are responsible for securing any permissions needed for the reuse of copyrighted materials included in the manuscript.




